BS5002 - Cellular Biochemistry (TERM 2 | WEEK 1 - Cloning a disease gene)
Key learning objectives list:
- LO1 - How to prepare 'donor DNA' for cloning
- LO2 - Understand the concept of gene libraries
- LO3 - Understand phage vectors used to create gene libraries
LO1 - How to prepare 'donor DNA' for cloning
Step 1: Choose a tissue
The final product of transcription of DNA is mature mRNA. This type of mRNA has been fully spliced (introns excised and exons ligated), leaving only the coding sequence of the gene.
Mature (spliced) mRNA. This mRNA can be translated directly to synthesise proteins in the ribosome.
It is important to choose a tissue that expresses a fairly large level of mRNA from the gene of interest.
Step 2: RNA extraction
Whilst various methods exist, one of the most common methods for extracting total RNA from a biological sample is AGPC (acid guanidinium thiocyanate-phenol-chloroform extraction).
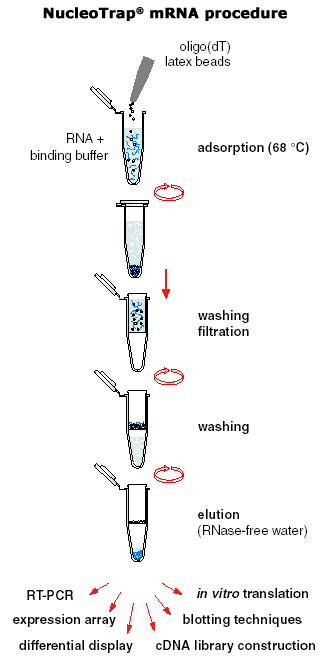
Step 3: mRNA purification
Again, there are numerous methods for mRNA isolation from total RNA, but the most common is affinity chromatography - a column purification method using magnetic beads within the matrix to attract the polyA tail.
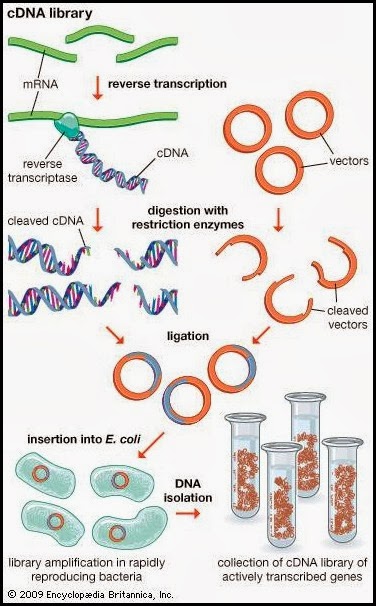
Step 4: Synthesise cDNA from mRNA
Using this mRNA and an enzyme called reverse transcriptase, we are able to create cDNA (complementary DNA).
LO2 - Understand the concept of gene libraries
cDNA Library vs. Genomic DNA Library - what is the difference?
cDNA libraries lack the non-coding and regulatory elements found in genomic DNA - useful for deducing protein coding sequences. cDNA libraries can be used to express eukaryotic genes in prokaryotes.
Genomic DNA libraries provide more detailed information about the organism, but are more resource-intensive to generate and maintain.
cDNA libraries lack the non-coding and regulatory elements found in genomic DNA - useful for deducing protein coding sequences. cDNA libraries can be used to express eukaryotic genes in prokaryotes.
Genomic DNA libraries provide more detailed information about the organism, but are more resource-intensive to generate and maintain.
LO3 - Understand phages used to create cloning vectors
Whilst plasmids are the most commonly used vectors for cloning, there is a limitation in the size of DNA that can be introduced to the cell via transformation.
Bacteriophage are a type of virus that can infect and replicate within bacteria. These can be modified into vectors that will transfer larger fragments of DNA.
- cDNA libraries are usually cloned into plasmid vectors.
- Genomic DNA libraries are usually cloned in bacteriophages
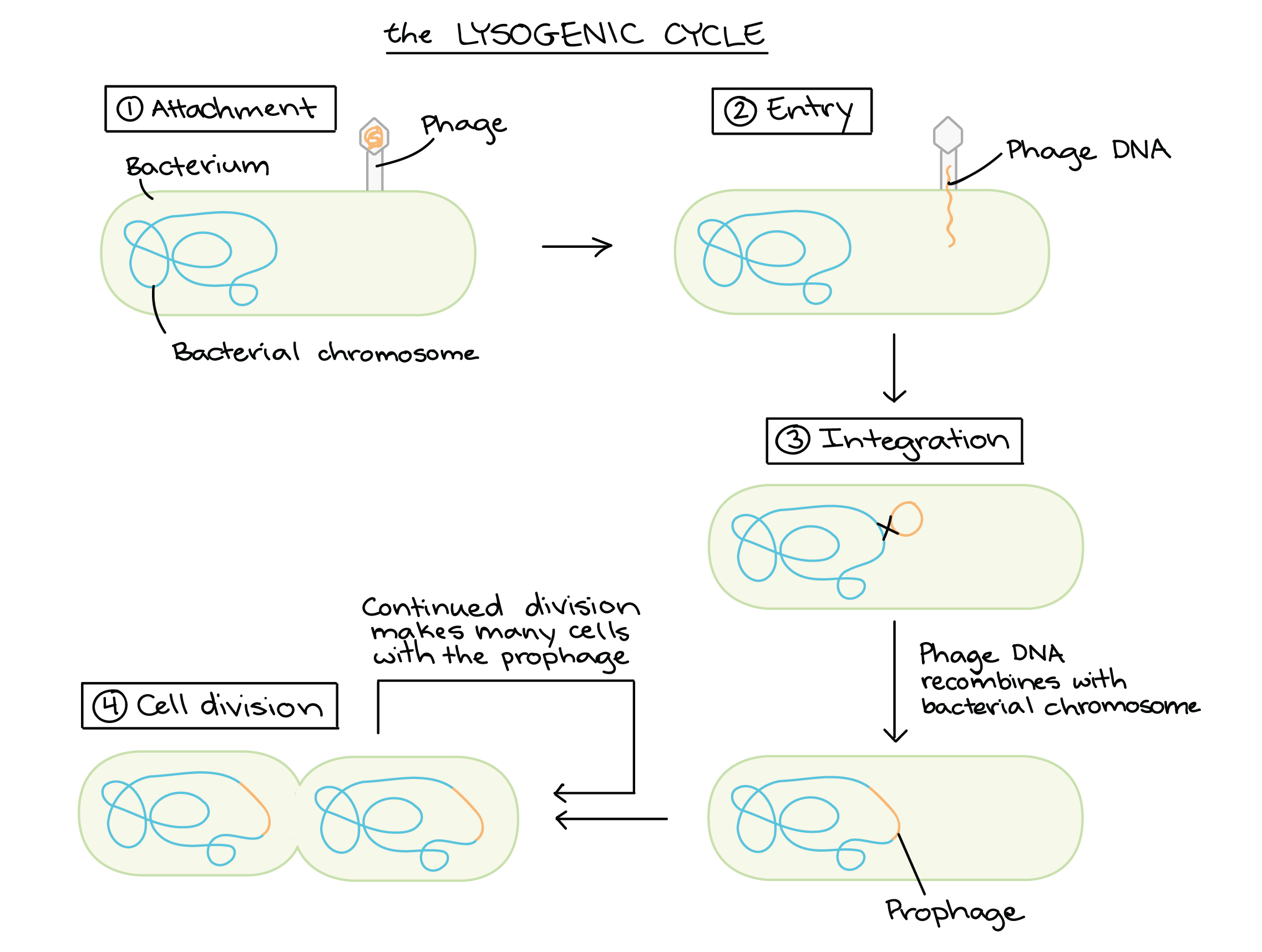
Phage λ
The Lysogenic Cycle
Now the λ bacteriophage has been prepared, it can be transferred into E-coli.
Plaque hybridization
1) Phage chromosome cleavage performed by EcoR1, a restriction endonuclease (or restriction enzyme) that cleaves double stranded DNA at the sequence GTTAAC. This allows for non-essential genes to be removed.
2) Complementary left arms are mixed with the foreign DNA we want to reproduce (which has also been cleaved by EcoRI)
3) Complementary 'stick ends' of the foreign DNA anneal to the native phage DNA
4) This 'recombinant' chromosome is now packaged into a λ protein coat
2) Complementary left arms are mixed with the foreign DNA we want to reproduce (which has also been cleaved by EcoRI)
3) Complementary 'stick ends' of the foreign DNA anneal to the native phage DNA
4) This 'recombinant' chromosome is now packaged into a λ protein coat
The Lysogenic Cycle
Now the λ bacteriophage has been prepared, it can be transferred into E-coli.
The 5 stages of the lysogenic cycle
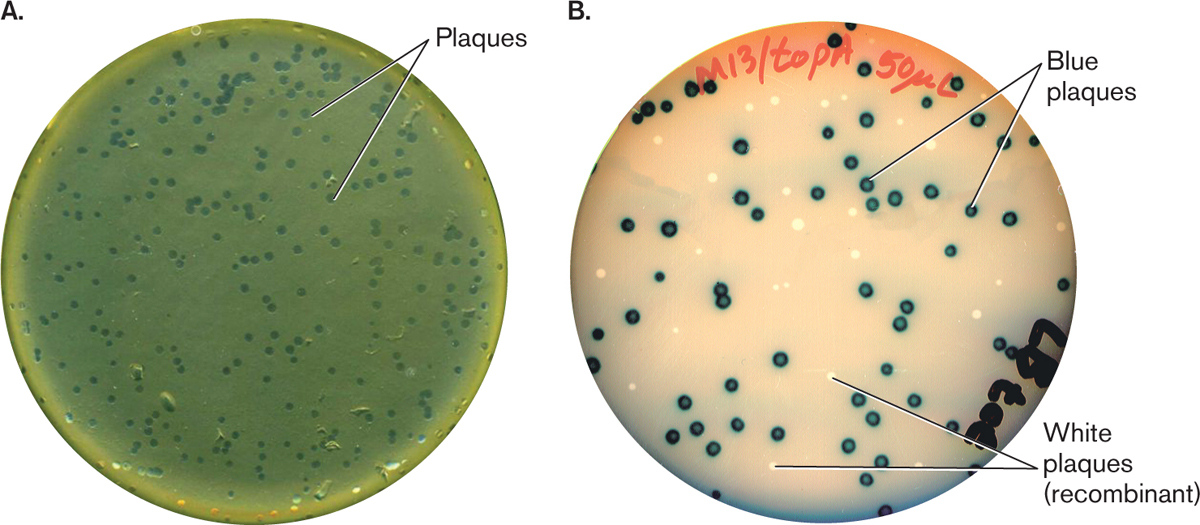
The bacteriophage are typically transferred into a 'lawn' of bacteria (usually E.coli). As the λ phage propagate, lysis of the bacteria will cause plaque formation.
A: Plaques formed by recombinant λ phage in E.coli. B: Plaques formed by recombinant M13 phage on E.coli
How do we find a gene within a gene library?
Now that we have created a genomic library on our plate, how do we find the gene of interest within it?
Large target DNA has been fragmented into individual genes and recombined with λ phage DNA. So how do we find the green gene, for example?








Comments
Post a Comment